출처: Grouped, stacked and percent stacked barplot in ggplot2 – the R Graph Gallery (r-graph-gallery.com)
Grouped barchart
A grouped barplot display a numeric value for a set of entities split in groups and subgroups. Before trying to build one, check how to make a basic barplot with R and ggplot2.
A few explanation about the code below:
- input dataset must provide 3 columns: the numeric value (value), and 2 categorical variables for the group (specie) and the subgroup (condition) levels.
- in the aes() call, x is the group (specie), and the subgroup (condition) is given to the fill argument.
- in the geom_bar() call, position="dodge" must be specified to have the bars one beside each other.
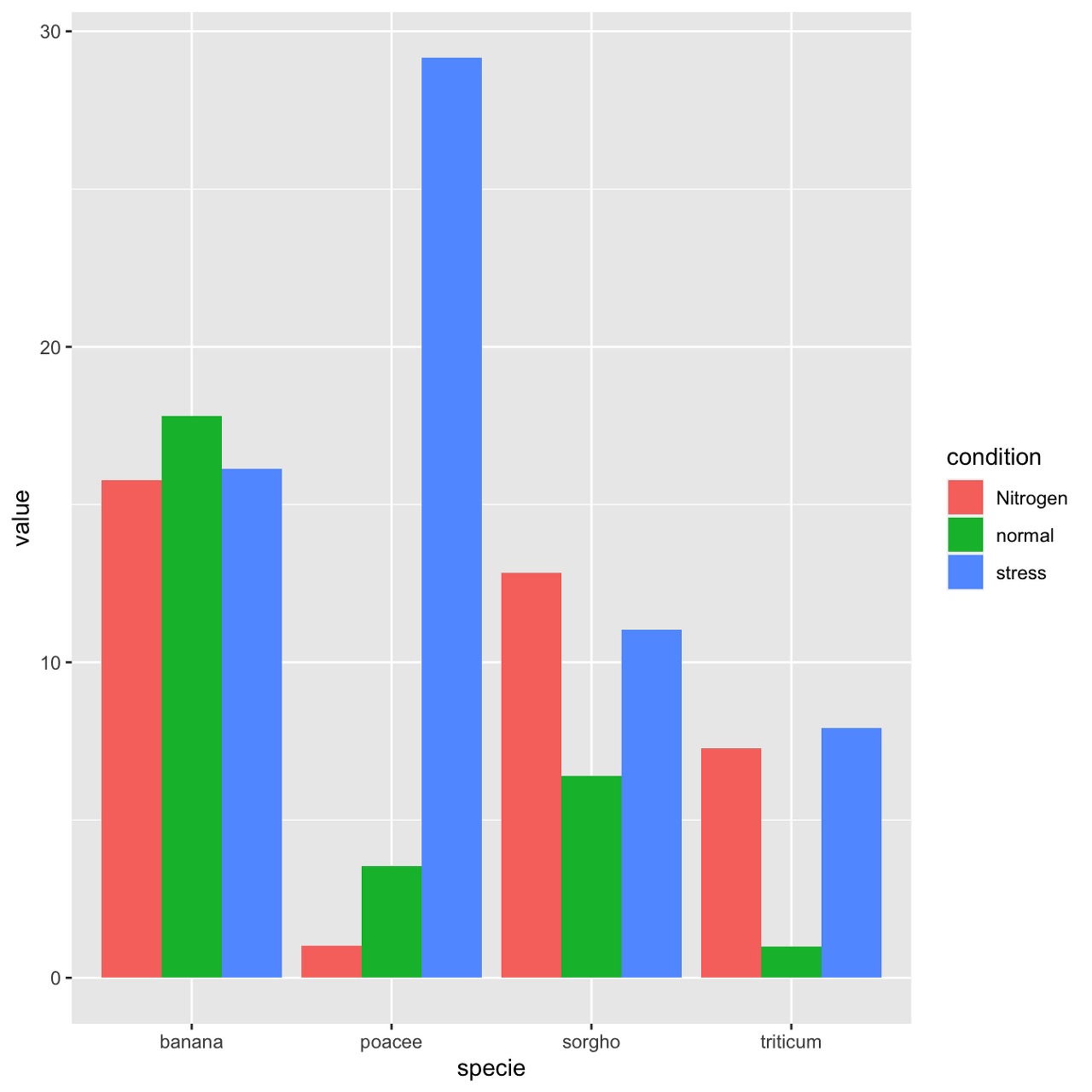
# library library(ggplot2) # create a dataset specie <- c(rep("sorgho" , 3) , rep("poacee" , 3) , rep("banana" , 3) , rep("triticum" , 3) ) condition <- rep(c("normal" , "stress" , "Nitrogen") , 4) value <- abs(rnorm(12 , 0 , 15)) data <- data.frame(specie,condition,value) # Grouped ggplot(data, aes(fill=condition, y=value, x=specie)) + geom_bar(position="dodge", stat="identity")
Stacked barchart
A stacked barplot is very similar to the grouped barplot above. The subgroups are just displayed on top of each other, not beside.
The only thing to change to get this figure is to switch the position argument to stack.

# library library(ggplot2) # create a dataset specie <- c(rep("sorgho" , 3) , rep("poacee" , 3) , rep("banana" , 3) , rep("triticum" , 3) ) condition <- rep(c("normal" , "stress" , "Nitrogen") , 4) value <- abs(rnorm(12 , 0 , 15)) data <- data.frame(specie,condition,value) # Stacked ggplot(data, aes(fill=condition, y=value, x=specie)) + geom_bar(position="stack", stat="identity")
Percent stacked barchart
Once more, there is not much to do to switch to a percent stacked barplot. Just switch to position="fill". Now, the percentage of each subgroup is represented, allowing to study the evolution of their proportion in the whole.
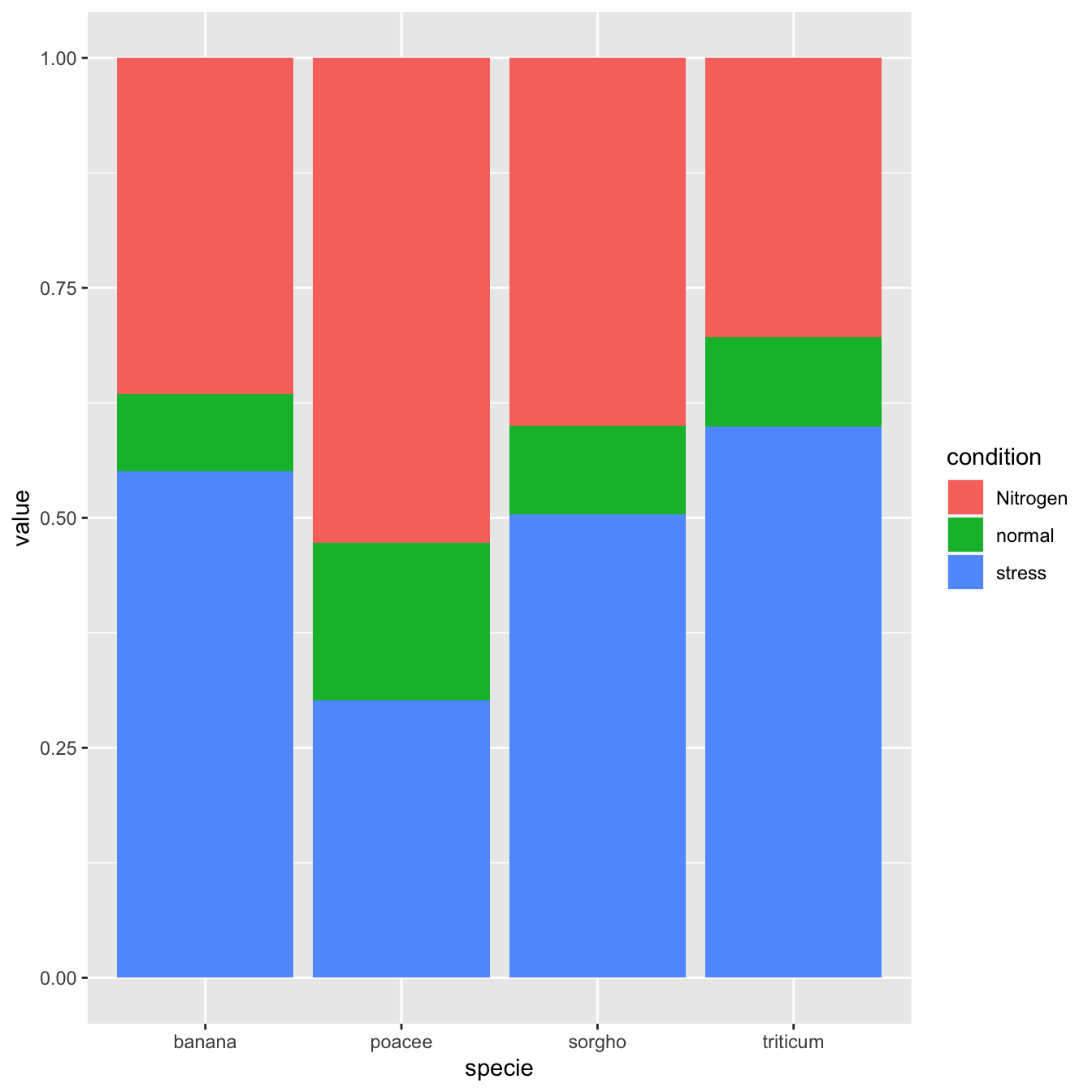
# library library(ggplot2) # create a dataset specie <- c(rep("sorgho" , 3) , rep("poacee" , 3) , rep("banana" , 3) , rep("triticum" , 3) ) condition <- rep(c("normal" , "stress" , "Nitrogen") , 4) value <- abs(rnorm(12 , 0 , 15)) data <- data.frame(specie,condition,value) # Stacked + percent ggplot(data, aes(fill=condition, y=value, x=specie)) + geom_bar(position="fill", stat="identity")
Grouped barchart customization
As usual, some customization are often necessary to make the chart look better and personnal. Let’s:

# library library(ggplot2) library(viridis) library(hrbrthemes) # create a dataset specie <- c(rep("sorgho" , 3) , rep("poacee" , 3) , rep("banana" , 3) , rep("triticum" , 3) ) condition <- rep(c("normal" , "stress" , "Nitrogen") , 4) value <- abs(rnorm(12 , 0 , 15)) data <- data.frame(specie,condition,value) # Small multiple ggplot(data, aes(fill=condition, y=value, x=specie)) + geom_bar(position="stack", stat="identity") + scale_fill_viridis(discrete = T) + ggtitle("Studying 4 species..") + theme_ipsum() + xlab("")
Small multiple
Small multiple can be used as an alternative of stacking or grouping. It is straightforward to make thanks to the facet_wrap() function.

# library library(ggplot2) library(viridis) library(hrbrthemes) # create a dataset specie <- c(rep("sorgho" , 3) , rep("poacee" , 3) , rep("banana" , 3) , rep("triticum" , 3) ) condition <- rep(c("normal" , "stress" , "Nitrogen") , 4) value <- abs(rnorm(12 , 0 , 15)) data <- data.frame(specie,condition,value) # Graph ggplot(data, aes(fill=condition, y=value, x=condition)) + geom_bar(position="dodge", stat="identity") + scale_fill_viridis(discrete = T, option = "E") + ggtitle("Studying 4 species..") + facet_wrap(~specie) + theme_ipsum() + theme(legend.position="none") + xlab("")